Transgenic mice
Transgenic mice
What are targeted transgenic mice?
The targeted transgenic mouse approach uses gene targeting by means of homologous recombination in embryonic stem (ES) cells. The transgene cDNA is knocked into a ‘safe habor’ locus in the mouse genome to generate transgenic mice with a predictable expression pattern without disturbing endogenous gene activity. Safe harbor loci have been utilised in the mouse to investigate gene function, generate humanized models and create tissue-specific reporter and Cre recombinase expressing lines.
What are targeted transgenic mice used for?
Targeted transgenic mouse models are developed by inserting the gene of interest into a specific region in the genome. This knock-in approach overcomes the limitation of random incorporation into the genome which can result in an undesirable expression level, spatiotemporal distribution of transgene activity, or even deleterious effects, thus limiting the usefulness of the model. Read our publications and research features for examples of targeted transgenics for more applications of this type of mouse model.

This technology has the following benefits:
- Only 1x transgenic mouse strain is needed, rather than multiple founder lines that may or may not express your gene of interest.
- Using only one mouse line saves time, money, resources, genotyping, phenotyping and animals.
- The 3Rs (refine, reduce, replace) can be achieved for the Institutional Animal Care and Use Committee (IACUC).
- Targeting into the safe habor locus achieves a ubiquitously inducible expression pattern.
- A predictable transgene-expressing animal is produced.
- The targeted transgenic strain has more utility, as it can be used for tissue-specific transgene expression simply by breeding to a different Cre mouse.
- Unwanted side effects caused by insertional mutagenesis associated with random transgenic mice are avoided.
- The targeted transgenic mice are readily bred to homozygosity.
- Data can be directly compared between targeted transgenic mouse lines. For example, various different point mutations can be compared in independent transgenic mice that were made or have been published.
How to make targeted transgenic mice?
Ozgene utilises either the Gt(ROSA)26Sor or Igs2 locus to generate targeted transgenic mice. Both ROSA26 and Igs2 are used as docking sites for transgenic constructs. Expression cassettes can be inserted within the locus so that they are expressed ubiquitously from an exogenous promoter.
Transgenic constructs are cloned into a shuttle vector and exchanged with the genomic sequence at the target locus via homologous recombination. Recombinant ES cells are then injected into goGermline™ mouse embryos to generate chimeras which can be bred to establish the required transgenic mouse model.
ROSA26 – Gt(ROSA)26Sor locus
The ROSA26 locus on chromosome 6 is the most commonly used site to generate inducible overexpression of the transgene because it does not contain any essential genes. The genomic structure of this locus is conserved between mice and humans. Expression cassettes can be inserted within the locus so that they are expressed ubiquitously from the endogenous ROSA26 promoter, or exogenous promoter elements can be introduced to drive transgene expression in a particular tissue type.
Igs2 locus
The Igs2 (H11, Hipp11) locus on chromosome 11 is an alternative safe harbor locus. Unlike the ROSA26 locus, Igs2 does not have an endogenous promoter. This makes targeting at the Igs2 locus highly flexible and particularly suitable for the conditional expression of transgenes engineered to be driven by a variety of tissue-specific promoters or otherwise inducible expression systems.
Conditional activation of gene insertions or substitutions
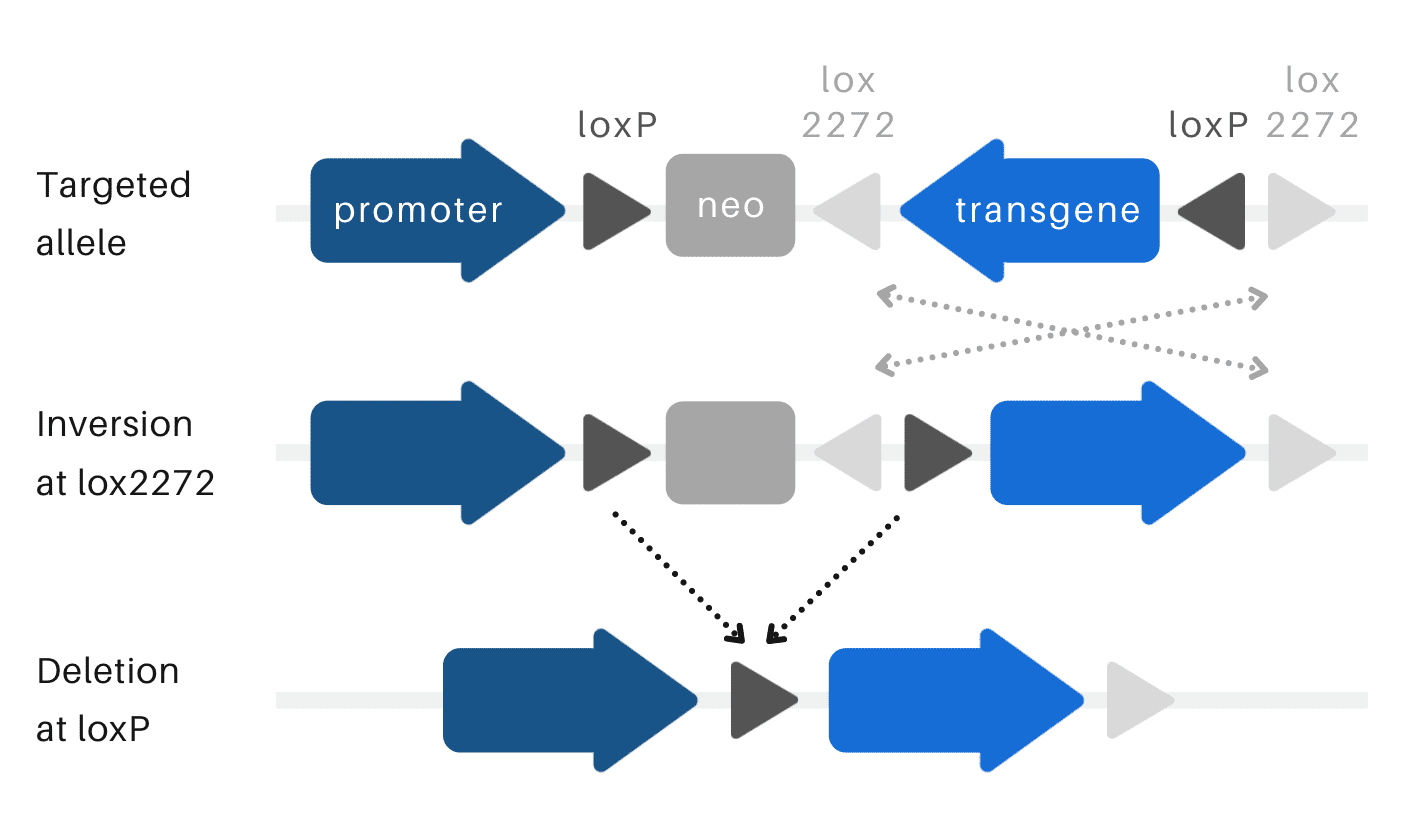
Ozgene uses a FLEx switch tool with two nested heterologous Lox sites in inverse orientation for the generation of conditional transgenic lines.

FLEx switch
Both LoxP and Lox2272 are recognised by Cre recombinase, but Lox2272 sites can only recombine with other Lox2272 sites, not with LoxP sites and vice versa. When the DNA sequence is flanked by Lox sites in opposing orientations, Cre will invert the sequence between the sites. If the DNA sequence is flanked by Lox sites in the same orientation, Cre will excise the sequence. The use of the FLEx switch results in both the deletion and inversion of a genetic sequence to activate the expression of the transgene in our targeted transgenic line.
How long does it take to generate mouse models?

“No project queues – get started today!”
Ozgene’s Lean management style coupled with patented goGermline technology allows us to generate mouse models in the fastest possible time together with increased certainty of germline transmission.
OzBIG – Genomic insertions up to 240 kb
Ozgene also offers a service called OzBIG for robust creation of large, gene-targeted replacement models. This system enables routine generation of humanized mice with very large (20 to 240 kb) genomic replacements and insertions. Even when the gene doesn’t exist in mice, OzBIG enables the expression of human genes by inserting them in the ROSA26 or Igs2 ‘safe harbor’ locus. OzBIG can also be used to generate large non-humanized, transgenics like complex reporters and synthetic expression systems.
Read more about Ozgene’s targeted transgenic
Intratumoural administration of an NKT cell agonist with CpG promotes NKT cell infiltration associated with an enhanced antitumour response and abscopal effect
Synthesis of human amyloid restricted to liver results in an Alzheimer disease-like neurodegenerative phenotype
The AGE receptor, OST48 drives podocyte foot process effacement and basement membrane expansion (alters structural composition)
Get in touch
We offer personalised services for your research needs. Request a free quote today.
Please fill out the form and we will respond to your query within two business days. Alternatively, visit our contact page for more ways that you can get in touch with us.
‘I received constant feedback, complete with gel images and experimental design. I felt I always knew what was going on with the project as if it were happening in my own lab.’
– Dr Maureen Neitz, University of Washington, USA


